Characterizing the performance of image segmentation approaches has been a persistent challenge. Performance analysis is important since segmentation algorithms often have limited accuracy and precision. Interactive drawing of the desired segmentation by human raters has often been the only acceptable approach, and yet suffers from intra-rater and inter-rater variability. Automated algorithms have been sought in order to remove the variability introduced by raters, but such algorithms must be assessed to ensure they are suitable for the task.
The performance of raters (human or algorithmic) generating segmentations of medical images has been difficult to quantify because of the difficulty of obtaining or estimating a known true segmentation for clinical data. Although physical and digital phantoms can be constructed for which ground truth is known or readily estimated, such phantoms do not fully reflect clinical images due to the difficulty of constructing phantoms which reproduce the full range of imaging characteristics and normal and pathological anatomical variability observed in clinical data.
Comparison to a collection of segmentations by raters is an attractive alternative since it can be carried out directly on the relevant clinical imaging data. However, the most appropriate measure or set of measures with which to compare such segmentations has not been clarified and several measures are used in practice. We developed an expectation-maximization algorithm for simultaneous truth and performance level estimation (STAPLE). The algorithm considers a collection of segmentations and computes a probabilistic estimate of the true segmentation and a measure of the performance level represented by each segmentation (Warfield, Zou, and Wells 2004).
The source of each segmentation in the collection may be an appropriately trained human rater or raters, or may be an automated segmentation algorithm. The probabilistic estimate of the true segmentation is formed by estimating an optimal combination of the segmentations, weighting each segmentation depending upon the estimated performance level, and incorporating a prior model for the spatial distribution of structures being segmented as well as spatial homogeneity constraints. STAPLE is straightforward to apply to clinical imaging data, it readily enables assessment of the performance of an automated image segmentation algorithm, and enables direct comparison of human rater and algorithm performance.
There has been significant interest in the STAPLE algorithm, and the Computational Radiology Laboratory at Children’s Hospital has provided the current implementation as open source software to research groups across the USA, as well as to interested scientists in Belgium, Switzerland, Canada, Israel, England, Sweden, France and Australia. The algorithm has been adopted and used by many collaborators at Children’s Hospital, collaborators across Boston, and also by national research efforts including NA-MIC and BIRN.
Software for Validation of Image Segmentation
There are currently relatively few tools available for providing validation of image segmentation. One example is ValMet available from the University of North Carolina, which implements a number of commonly used spatial overlap and distance measures, but which does not estimate a reference standard and does not implement the STAPLE algorithm.
Rohlfing and colleagues (Rohlfing, Russakoff, and Maurer 2004; Rohlfing et al. 2004) have demonstrated that the STAPLE algorithm enables optimal combinations of classifiers to be computed, and have shown this can significantly improve the results of segmentation by registration. In this scheme, several existing segmentations are aligned to a target image that is to be segmented, usually using nonrigid registration, and then an optimal combination of the projected segmentations is used to remove the impact of registration errors.
Illustration of Application
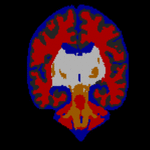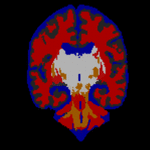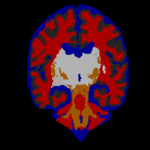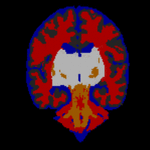
STAPLE was applied to the collection of segmentations, and a reference standard segmentation obtained for each subject. In order to readily visualize the probabilistic reference standard here, it was converted to a hard labeling by maximum likelihood estimation, and color coded. An illustrative MR scan of a newborn infant, interactive segmentations and reference standard estimate are illustrated in the figure below.
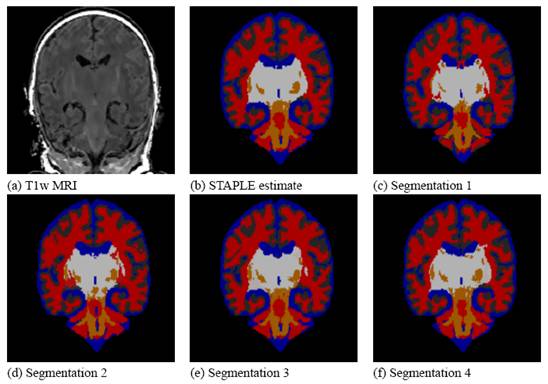
STAPLE computes a reference standard from a collection of segmentations
This presentation also illustrates some of the visualization difficulties intrinsic in displaying a probabilistic reference standard. Here we have converted the reference standard to a ``hard’’ labeling by maximum likelihood classification.
Recent Work
In (Warfield, Zou, and Wells 2008, 2006) we described a new algorithm to enable the estimation of performance characteristics, and a true labelling, from observations of segmentations of imaging data where segmentation labels may be ordered or continuous measures. This approach may be used with, among others, surface, distance transform or level-set representations of segmentations, and can be used to assess whether or not a rater consistently overestimates or underestimates the position of a boundary.
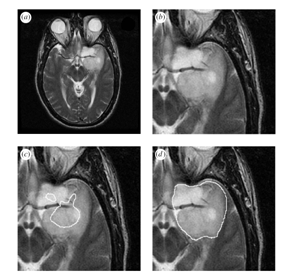
Average and estimated true tumour segmentations from human rater segmentations, binarized by thresholding the estimated true score at 0.0.
[a] MRI of a brain tumour. [b] Region of interest. [c] Binarized average of rater segmentations. [d] Binarized estimated tumour segmentation from new algorithm.
References
- S K Warfield, K H Zou, and W M Wells. 2004. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 23 (7):903-21. [WWW] [PDF]
- S K Warfield, K H Zou, and W M Wells. 2006. Validation of image segmentation by estimating rater bias and variance. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv 9 (Pt 2):839-47. [WWW]
- S K Warfield, K H Zou, and W M Wells. 2008. Validation of image segmentation by estimating rater bias and variance. Philos Transact A Math Phys Eng Sci 366 (1874):2361-75. [WWW] [PDF]